The Sci2 Tool is an 'empty shell' filled with plugins. Some plugins run on the core architecture, OSGi and CIShell (see Section 7 Extending the Sci2 Tool). Others convert loaded data into in-memory objects, formatted for different algorithms to read it. The algorithm plugins themselves can be divided into different menus, in this case data preparation, preprocessing, analysis, modeling and visualization. Users are not limited to using pre-packaged plugins; instead, they can create, download, share, and import their own.
There are two ways plugins can be added to the menu. First, if an algorithm is listed in the default_menu.xml file, which is found in the configuration/ directory of a CIShell tool, it is added in the place specified by the default_menu.xml file. Second, if an algorithm is not listed in this file, but has specified a menu_path property in the algorithm.properties file, the menu manager CIShell service will add it to the appropriate menu, as described in the by the menu_path property's value. For example, Analysis/additions will place an algorithm on the bottom of the Analysis menu.
It is possible to extend Sci2 Tool, by adding plugins related to database functionalities and Cytoscape tool (open source software platform for visualizing networks).
By adding the plugins related to database functionalities, the Sci2 Tool can support the creation of databases from ISI and NSF files. Database loading improves the speed and functionality of data preparation and preprocessing. While the initial loading can take quite some time for larger datasets (see sections 3.4 Memory Allocation and 3.5 Memory Limits) it results in vastly faster and more powerful data processing and extraction.
Cytoscape (http://www.cytoscape.org) is an open source software platform for visualizing networks and integrating these with any type of attribute data. Although Cytoscape was originally designed for biological research, now it is a general platform for network analysis and visualization. A variety of layout algorithms are available, including cyclic, tree, force-directed, edge-weight, and yFiles Organic layouts. Cytoscape tool can also be added to the Sci2 Tool as a plugin, permitting that the networks generated at Sci2 be visualized in Cystoscape.
To add Database, Cytoscape, Congressional District Geocoder, and New ISI File Format plugins to Sci2
- Download these files that contain additional plugins to add Database, Balloon Graph, Cytoscape, Congressional District Geocoder, and New ISI File Format plugins to Sci2:
- Sci2 v. 0.5.2 beta or older
- Sci2 Database plugins - DatabasePlugins.zip (unzip to obtain the jar files that will be inside a folder named DatabasePlugins) - Note the Database plugins should only be used with Sci2 v0.5.2 alpha or older
- ISI Web of Science file format - WOS-plugins.zip - plugin provides the support for the newest version of the Web of Science data format, see ISI (*.isi) for more information
- Sci2 v. 1.0 beta or older
- UCSD Map of Science - edu.iu.sci2.visualization.scimaps_0.1.0.jar - Note this plugin is for should only be used with Sci2 v1.0 beta or older
- Update Temporal Bar Graph for European dates - edu.iu.sci2.visualization.temporalbargraph_0.0.10.jar - plugin fixes the temporal bar graph plugin to work with European date format.
- Update Extract Word Co-Occurrence Network - edu.iu.nwb.composite.extractcowordfromtable_1.0.1.jar - plugin fix creates isolated nodes for unique record ID
- Twitter Reader - edu.iu.sci2.reader.twitter_1.0.0.jar pulls information from Twitter. For more information on how to use this algorithm, (currently not working) see Twitter Reader Documentation.
- Current Release or older
- Balloon Graph - BalloonGraph.zip (unzip to obtain the jar files that will be inside a folder named BalloonGraph)
- Cytoscape network visualization plugin - org.textrend.visualization.cytoscape_0.0.3.jar
- Congressional District Geocoder
- Most recent version, for 113th Congress: edu.iu.sci2.preprocessing.zip2district_0.0.2.jar
- Deprecated version, for 112th Congress: edu.iu.sci2.preprocessing.zip2district_0.0.1.jar
- Most recent version, for 113th Congress: edu.iu.sci2.preprocessing.zip2district_0.0.2.jar
- Sci2 v. 0.5.2 beta or older
- You will have to extract these files before you copy them into plugins directory in the Sci2 folder
- Open the Sci2 folder and go to the plugins directory:
- Copy the plugin files themselves (not the folders that contain them) into the directory /plugins of Sci2 directory:
- Download the file default_menu.xml by right-clicking and saving the link as an XML file. Then copy this file into the directory configuration/ under Sci2 directory, replacing the older version of this file. This file specifies where each plugin added will be located at the menu. Note, this step only applies if you have downloaded the database plugins for Sci2 v0.5.2 alpha or older.
- The next time the Sci2 Tool is loaded after this modifications, these additional plugins will appear as new menu items.
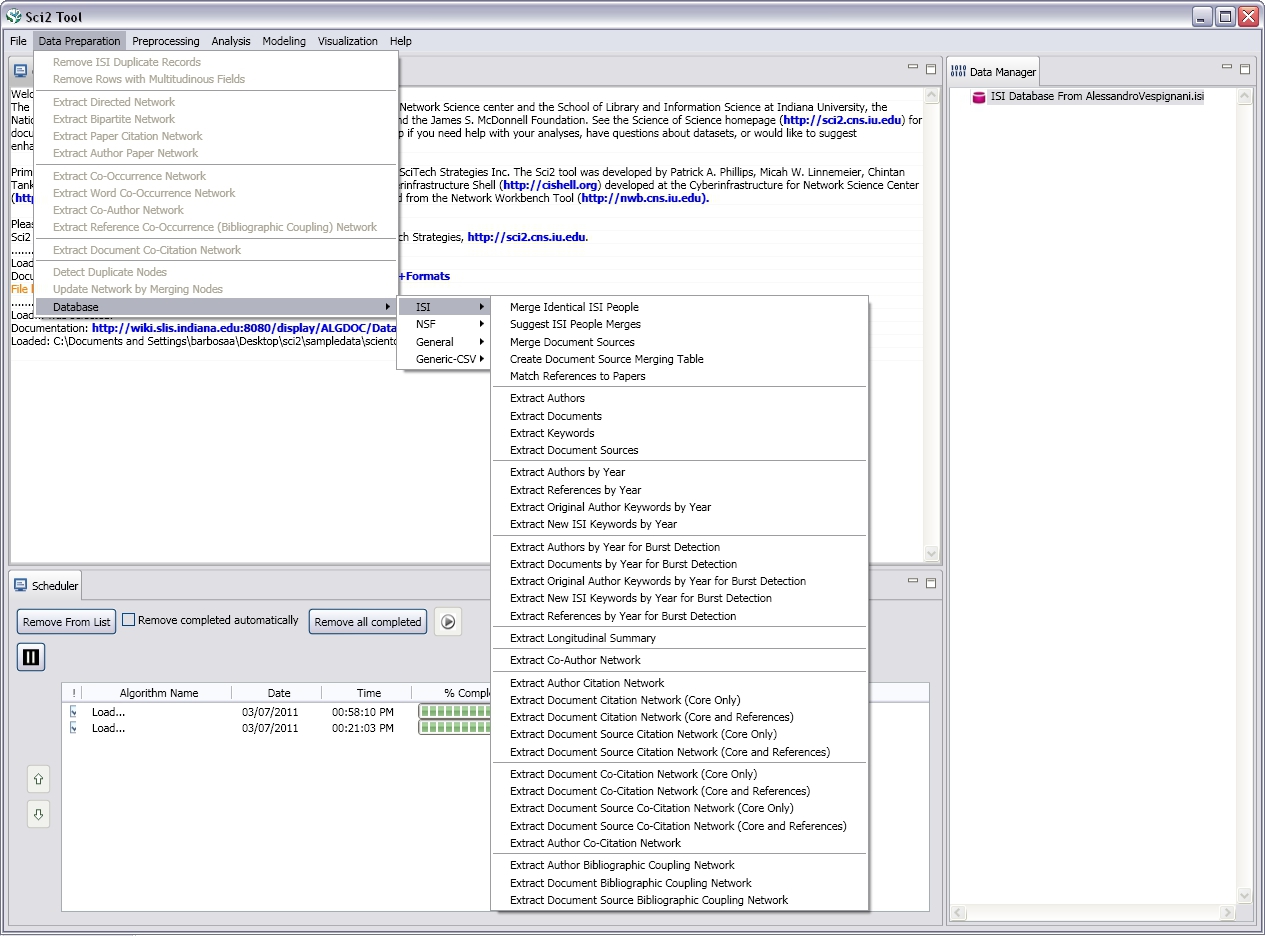
Algorithms available at extended version
Database
ISI
- Merge Identical ISI People – Merges all people in an ISI database who have the same name.
- Suggest ISI People Merges – Generates a pre-annotated merging table based on a user-selected threshold and string similarity metric.
- Merge Document Sources – Merge document sources of documents and references based upon known name variants and abbreviations.
- Create Document Source Merging Table – Essentially the same as Create Merging Table, but including two analyses of your dataset that may help in determining good candidates for merging.
- Match References to Papers – Matches references to papers if they have the same first author, source (journal), start page, volume, and year.
--------------------------------------------- - Extract Authors – Outputs a table containing one row per author in the database.
- Extract Documents – Outputs a table containing one row per document in the database.
- Extract Keywords – Outputs a table containing one row per keyword in the database.
- Extract Document Sources – Outputs a table containing one row per document source in the database.
--------------------------------------------- - Extract Authors by Year – Outputs a table containing the amount of publications per author per year.
- Extract References by Year – Outputs a table containing the amount of references to a publication per year.
- Extract Original Author Keywords by Year – Outputs a table containing one row per original author keyword per year.
- Extract New ISI Keywords by Year – Outputs a table containing one row per new ISI keyword per year.
--------------------------------------------- - Extract Authors by Year for Burst Detection – Used for author burst detection.
- Extract Documents by Year for Burst Detection – Used for word occurrence based burst detection.
- Extract Original Author Keywords by Year for Burst Detection – Used for author keyword burst detection.
- Extract New ISI Keywords by Year for Burst Detection – Used for new ISI keyword burst detection.
- Extract References by Year for Burst Detection – Used to detecting bursting references to publications.
--------------------------------------------- - Extract Longitudinal Summary – Outputs a table with the total number of documents published, references published, references made, distinct authors, distinct sources, distinct author keywords, distinct ISI keywords, and distinct other keywords by year.
--------------------------------------------- - Extract Co-Author Network – Extracts a weighted, undirected network with authors as nodes and edges between authors who co-wrote papers. The extraction appends to nodes the number of authored documents, ISI's times-cited count, the publication of the earliest document, and the publication year of the most recent document. The extraction appends to edges weights for the number of co-written papers, and the publication years of the earliest and most recent collaboration.
--------------------------------------------- - Extract Author Citation Network – Extracts a weighted, directed network with authors as nodes and edges from a citing author to a cited author. Nodes include all data from the PERSON table and the number of documents authored in the current dataset.
- Extract Document Citation Network (Core Only) – Extracts an unweighted, directed network with documents as nodes and edges from a citing paper to a cited paper. Only those documents with full entries in the dataset are included in the network. Nodes include all data from the DOCUMENT table.
- Extract Document Citation Network (Core and References) – Extracts an unweighted, directed network with documents as nodes and edges from a citing paper to a cited paper. Nodes include all data from the DOCUMENT table.
- Extract Document Source Citation Network (Core Only) – Extracts a weighted, directed network with sources (journals) as nodes and edges from a citing source to a cited source. Citations are via documents within sources, and only those sources represented by documents within the dataset are included in the network. Nodes include all data from the SOURCE table, and edges are weighted by the number of citations between sources.
- Extract Document Source Citation Network (Core and References) – -- Extracts a weighted, directed network with sources (journals) as nodes and edges from a citing source to a cited source. Citations are via documents within sources. Nodes include all data from the SOURCE table, and edges are weighted by the number of citations between sources.
--------------------------------------------- - Extract Document Co-Citation Network (Core Only) – Extracts a weighted, undirected network with documents as nodes and edges between documents which have been cited together. Only those documents with entries in the dataset are included in the network. Edge weight is determined by the number of times two articles are cited together, and edges are appended with the publication years of their earliest and most recent co-citations.
- Extract Document Co-Citation Network (Core and References) – Extracts a weighted, undirected network with documents as nodes and edges between documents which have been cited together. Edge weight is determined by the number of times two articles are cited together, and edges are appended with the publication years of their earliest and most recent co-citations.
- Extract Document Source Co-Citation Network (Core Only) – Extracts a weighted, undirected network with journals as nodes and edges between journals which have been cited together by a common document. Only those journals containing documents with entries in the dataset are included in the network. Edge weight is determined by the number of times two journals are cited together, and edges are appended with the publication years of their earliest and most recent co-citations.
- Extract Document Source Co-Citation Network (Core and References) – Extracts a weighted, undirected network with journals as nodes and edges between journals which have been cited together by a common document. Edge weight is determined by the number of times two journals are cited together, and edges are appended with the publication years of their earliest and most recent co-citations.
- Extract Author Co-Citation Network – Extracts a weighted, undirected network with authors as nodes and edges between authors who have been cited together by a common document. Edge weight is determined by the number of times two authors are cited together, and edges are appended with the publication years of their earliest and most recent co-citations.
--------------------------------------------- - Extract Author Bibliographic Coupling Network – Extracts a weighted, undirected network with authors as nodes and edges between authors who cite a common reference. Edge weight is determined by the number of common references between authors.
- Extract Document Bibliographic Coupling Network – Extracts a weighted, undirected network with documents as nodes and edges between documents which cite a common reference. Edge weight is determined by the number of common references between documents.
- Extract Document Source Bibliographic Coupling Network – Extracts a weighted, undirected network with journals as nodes and edges between journals whose documents cite a common reference. Edge weight is determined by the number of common references between documents.
NSF
- Merge Identical NSF People – Merges all people in an NSF database who have the same name.
--------------------------------------------- - Extract Investigators – Extracts a table containing one row per investigator from an NSF database.
- Extract Awards – Extracts a table containing one row per award from an NSF database.
- Extract Organizations – Extracts a table containing one row per organization from an NSF database.
--------------------------------------------- - Extract Co-PI Network – Extracts a weighted, undirected network with principle investigators as nodes and edges between them if they co-investigated an award in the database. Nodes are appended with the number of awards investigated, total amount across each investigated award, start date of earliest award, and expiration date of most recent award. Edges are appended with the number of awards co-investigated by the two investigators and the joint award total between the investigators.
- Merge Identical NSF People – Merges all people in an NSF database who have the same name.
General
- Create Merging Table – Given a database, this generates a spreadsheet that can be annotated to indicate how entities should be merged. The user selects the type of entity.
- Merge Entities – Given a database and a merging table, this generates a new database where the merges in the merging table have been performed.
- Custom Table Query – This algorithm executes the given SQL query on the selected database, returning a table with the results.
- Custom Graph Query – This algorithm executes the two given SQL queries on the selected database, returning a graph with the results.
- Extract Raw Tables from Database – Lets you explore the raw contents of a database, extracting every table in the database into the Data Manager.
Generic-CSV
- Extract Co-Occurrence Network – Allows the user to create a co-occurrence network based on columns in a table.
- Extract Bipartite Network – Extract a bipartite network from tabular data.
- Extract Co-Entity Occurrence Network
- Extract Table
Analysis
Geospatial
- Congressional District Geocoder - This algorithm converts the given 9-digits U.S. ZIP codes (ZIP+4 codes) into its congressional districts and geographical coordinates (latitude and longitude).
Visualization
Networks
- Cytoscape - Open source software platform for visualizing networks and integrating these with any type of attribute data.
- Bipartite Network Graph
Menus at Extended Version
As stated before, this additional plugins will appear as new menu items in the Sci2 Tool.
Figure 2.1: Menus in the extended version of Sci2.
How Algorithms are Added to the Menu
There are two ways algorithms are added to the menu. First, if an algorithm is listed in the default_menu.xml file, which is found in the configuration/ directory of the Sci2 Tool, it is added in the place specified by the default_menu.xml file. Secondly, if an algorithm is not listed in this file, but has specified a menu_path property in the algorithm.properties file, the menu manager CIShell service will add it to the appropriate menu, as described in the by the menu_path property's value (for example, Analysis/additions will place an algorithm on the bottom of the Analysis menu).